ARCIMBOLDO_SHREDDER Predicted Models tutorial
Aims of the tutorial
This tutorial shows how to launch the predicted_model mode1 implemented in ARCIMBOLDO_SHREDDER2, which has been developed to optimize the use of predicted models, while systematically removing model bias.
The predicted_model mode will automatically pre-process the predicted models (either from AF2 or RoseTTAFold), decompose them into structural units considering domains, and use them for phasing. For very partial solutions, successful expansion will suffice as verification since errors in the starting model prevent phase improvement. Straightforward solutions are verified by systematically removing model bias using SHELXE3, which will omit the original fragment from the trace. Subsequently, all consistent traces are combined in reciprocal space with ALIXE4. In the case of coiled-coils5 the verification will be performed by scoring the best solution against a baseline complying with the modulation in the data. Finally, if the structure is a multimer and expansion of a first placement fails to yield a solution, the multicopy1 procedure will be activated to search further copies sequentially.
This example uses the latest ARCIMBOLDO_SHREDDER as of August 2023.
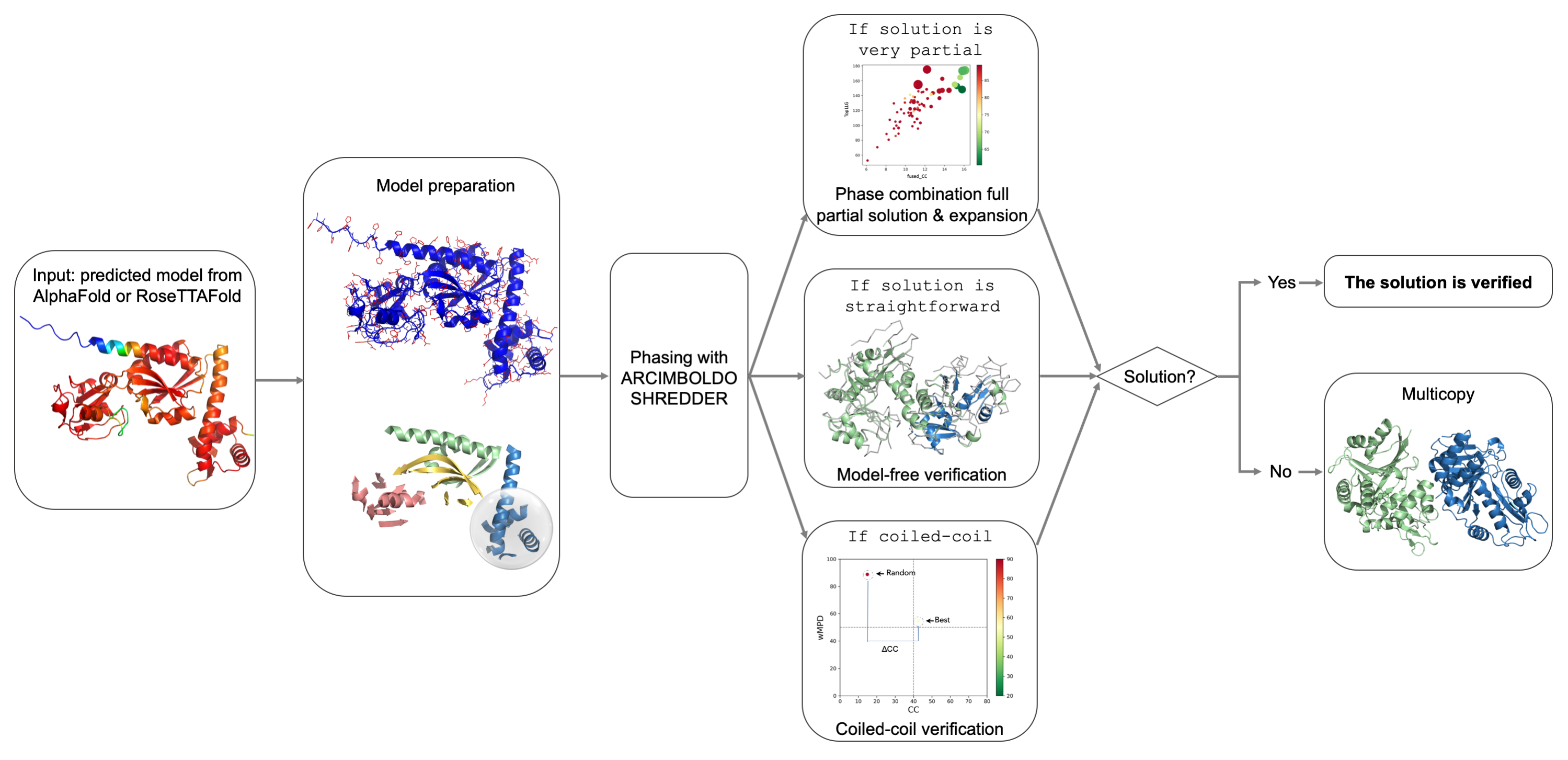
Figure 1. The predicted_model mode from ARCIMBOLDO_SHREDDER workflow.
Experimental details and data
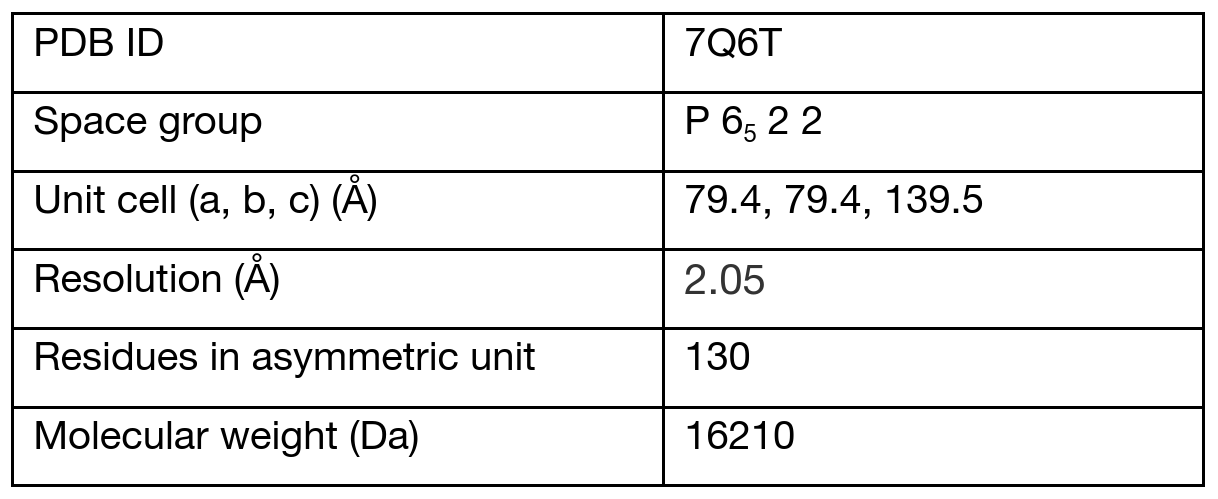
Test data for our tutorial is the crystal structure of the bromo-domain of ATAD2 with AZ1382437. The structure was solved in 2021 by molecular replacement and is deposited in the Protein Data Bank under the PDB code 7Q6T6.
Details of the data are summarized in the following table:
AlphaFold models were generated on the cloud through the Google Colaboratory Notebook MMseqs27.
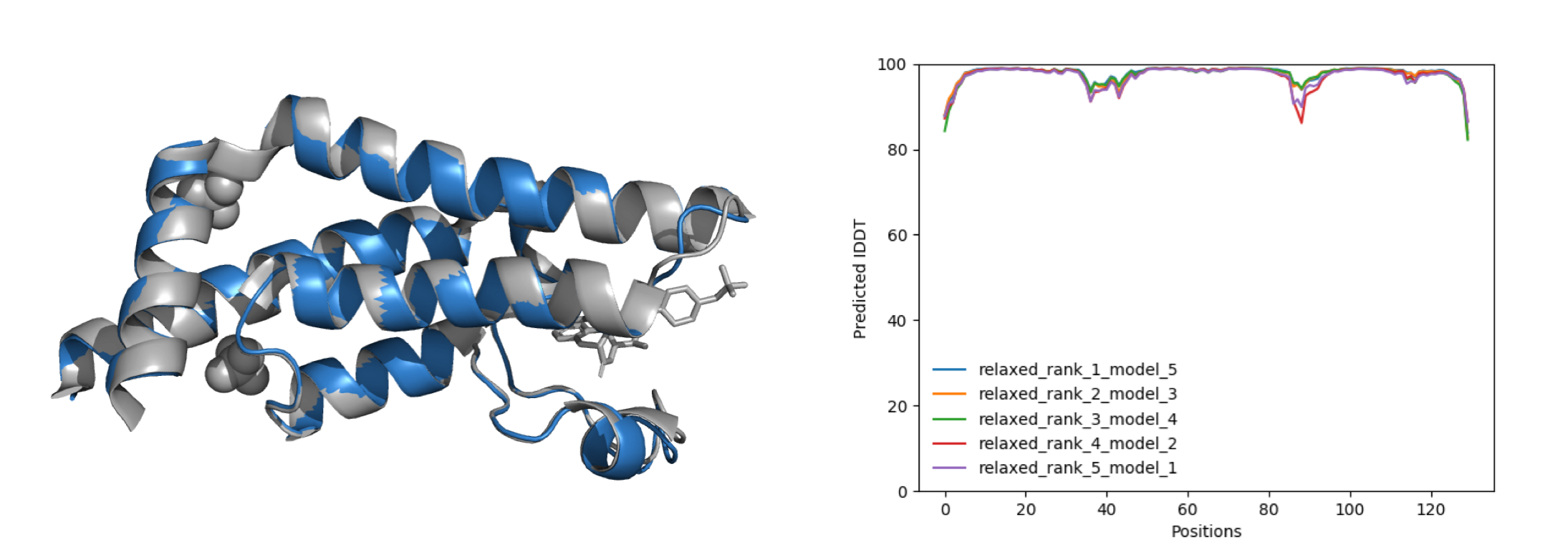
Figure 2. Superposition of the best-ranked model (blue) and the final structure of PDB entry 7Q6T (grey) (left) and quality of the models represented by pLDDT in Å per position (right).
Step by Step tutorial
Input
All required files can be downloaded here.
The description of the configuration file follows:
[CONNECTION]: distribute_computing: local_grid setup_bor_path: /path/to/setup.bor [LOCAL] path_local_phaser: /path/to/local_phaser path_local_shelxe: /path/to/local_shelxe [GENERAL]: working_directory: /path/to/working_directory mtz_path: %(working_directory)s/7q6t.mtz hkl_path: %(working_directory)s/7q6t.hkl [ARCIMBOLDO-SHREDDER] name_job: 7q6t molecular_weight: 16210 number_of_component: 1 f_label: F sigf_label: SIGF model_file: /absolute/path/to/7q6t_af_model.pdb rmsd_shredder: 0.8 shred_method: spherical predicted_model: True
In the .bor file you need to specify:
In the [ARCIMBOLDO-SHREDDER] section you will need to define the contents of the asymmetric unit, the labels for the mtz file and the path to the AlphaFold or RossetaFold model.
Another parameter that has default but that may be changed is the expected rmsd of the models (rmsd_shredder), in this case, we will change the default value from 1.2 to 0.8 Å. Finally, select the SHREDDER method to spherical (the other option is sequential, but spherical is the default and in which the predicted_model mode works), and set the predicted_model mode to True.
Execution
Terminal
You can run the program from a terminal, redirecting the output to a log file.
1. Interactively
ARCIMBOLDO_SHREDDER 7q6t.bor
2. In background
nohup ARCIMBOLDO_SHREDDER 7q6t.bor >& log &
CCP4
The predicted_model mode of ARCIMBOLDO_SHREDDER can run in CCP4i, CCP4i2, and CCP4 online8. In this case .bor file is generated within the interfaces. Follow the detailed instructions here.Output and Results
The HTML output file summarizes your job instructions and results. This file is updated during the run and can be opened in an internet browser.
In the directory where you launched ARCIMBOLDO_SHREDDER, you will find a directory with a library of equal-sized spherical fragments of comparable scattering generated from the predicted model. Then, the directory called ARCIMBOLDO_BORGES contains the output of the ARCIMBOLDO_BORGES run using this library of models.
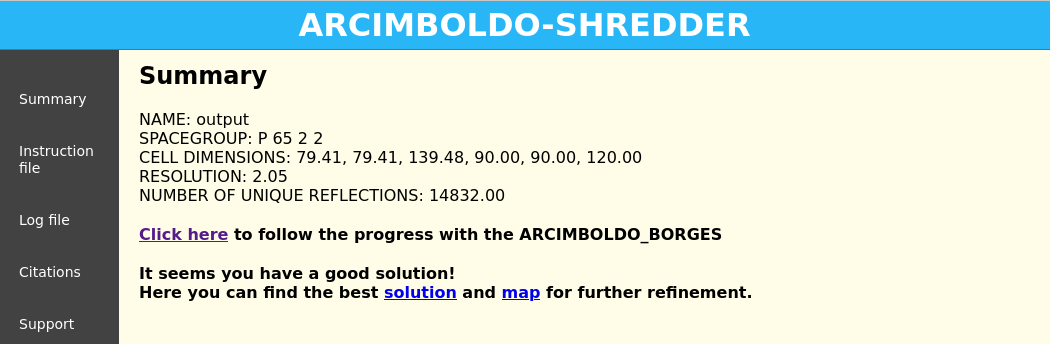
Figure 3. Click on “Click here” to access the ARCIMBOLDO_BORGES results.
The first section is a summary of the parameters of the structure. The next section displays a graph and a table summarizing the rotation clustering step as in ARCIMBOLDO_BORGES. A sortable table follows, summarizing the results for all PHASER and SHELXE steps, including top and average figures of merit for each rotation cluster that has been evaluated. Then the backtracking section contains links to access the best current solution: the pdb of the traced structure and the map in .phs format. The last section of the HTML echoes all parameters used for the run so that defaults are listed along with values for the parameters set through the .bor file. This allows the user to reproduce the run even if defaults may change in future versions.
Figure 4. Backtracking of the best current solution. A correlation coefficient of 61% with 128 residues traced is obtained. Numerical values may vary slightly.
References
-
Verification: model-free phasing with enhanced predicted models in ARCIMBOLDO_SHREDDER.
Medina, A., Jiménez, E., Caballero, I., Castellví, A., Triviño Valls, J., Alcorlo, M., Molina, R., Hermoso, J. A., Sammito, M. D., Borges, R. & Usón, I.
Acta Cryst. D78, 1283–1293 (2022) (doi:10.1107/S2059798322009706)
-
Exploiting distant homologues for phasing through the generation of compact fragments, local fold refinement and partial solution combination.
Millán, C., Sammito, M. D., McCoy, A. J., Nascimento, A. F., Petrillo, G., Oeffner, R. D., Domínguez-Gil, T., Hermoso, J. A., Read, R. J. and Usón, I.
Acta Cryst. D74: 290-304 (2018) (doi:10.1107/S2059798318001365)
-
An introduction to experimental phasing of macromolecules illustrated by SHELX; new autotracing features.
Uson, I. and Sheldrick, G.M.
Acta Cryst. D74, 106-116 (2018) (doi:10.1107/S2059798317015121)
-
ALIXE: a phase-combination tool for fragment-based molecular replacement.
Millán, C., Jiménez, E., Schuster, A., Diederichs, K. & Usón, I.
Acta Cryst. D76, 209–220 (2020) (doi:10.1107/S205979832000056X)
-
ARCIMBOLDO on coiled coils.
Caballero, I., Sammito, M., Millan, C., Lebedev, A., Soler, N. and Uson, I.
Acta Cryst. D74, 194-204 (2018) (doi:10.1107/S2059798317017582)
-
Discovery of a Potent and Selective ATAD2 Bromodomain Inhibitor with Antiproliferative Activity in Breast Cancer Models.
Winter-Holt, J. J., Bardelle, C., Chiarparin, E., Dale, I. L., Davey, P. R. J., Davies, N. L., Denz, C., Fillery, S. M., Guérot, C. M., Han, F., Hughes, S. J., Kulkarni, M., Liu, Z., Milbradt, A., Moss, T. A., Niu, H., Patel, J., Rabow, A. A., Schimpl, M., Shi, J., Sun, D., Yang, D. & Guichard, S.
J. Med. Chem. 65, 3306–3331 (2022) (doi:10.1021/acs.jmedchem.1c01871)
-
ColabFold: making protein folding accessible to all.
Mirdita, M., Schütze, K., Moriwaki, Y., Heo, L., Ovchinnikov, S. & Steinegger, M.
Nat. Methods, 19, 679–682 (2022) (doi:10.1038/s41592-022-01488-1)
-
Predicted models and CCP4.
Simpkin, A.J., Caballero, I., McNicholas, S., Stevenson, K., Jiménez, E., Sánchez Rodríguez, F., Fando, M., Uski, V., Ballard, C., Chojnowski, G., Lebedev, A., Krissinel, E., Usón, I., Rigden, D.J., Keegan, R.M.
Acta Cryst. (2023) In press