ARCIMBOLDO on DNA
ARCIMBOLDO on DNA: Structure solution of DNA-binding proteins and complexes with ARCIMBOLDO libraries
The crystallographic particularities of nucleic acids tend
to pose specific challenges to methods primarily developed
for proteins. Crystallographic structure solution of protein-DNA
complexes therefore remains a challenging area that is in need of optimized
experimental and computational methods.
The potential of
the structure-solution program ARCIMBOLDO for the
solution of protein-DNA complexes has therefore been
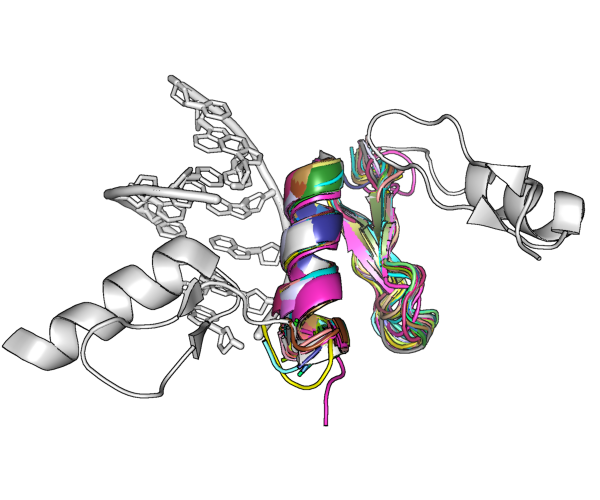
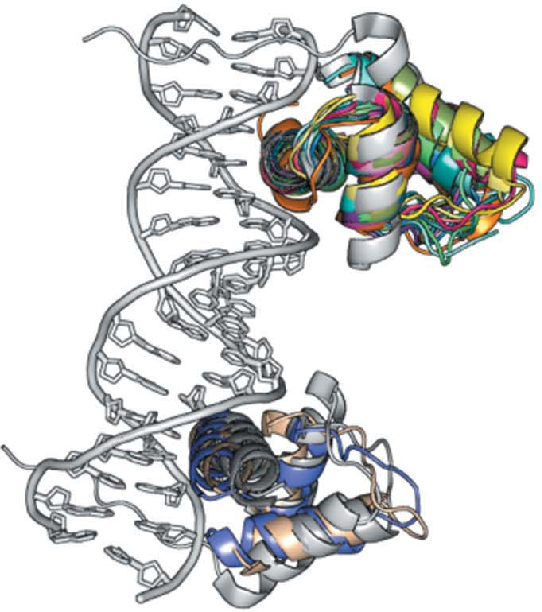
assessed. The method is based on the combination of locating
small, very accurate fragments using the program Phaser and
density modification with the program SHELXE.
Whereas for
typical proteins main-chain helices provide the ideal, almost
ubiquitous, small fragments to start searches, in the case of
DNA complexes the binding motifs and DNA double helix
constitute suitable search fragments.
Here
you can find instructions on how to solve protein-DNA complexes with ARCIMBOLDO_BORGES and how to create a library for one of the protein-DNA complexes shown on the right, or alternatively download a prepared library.
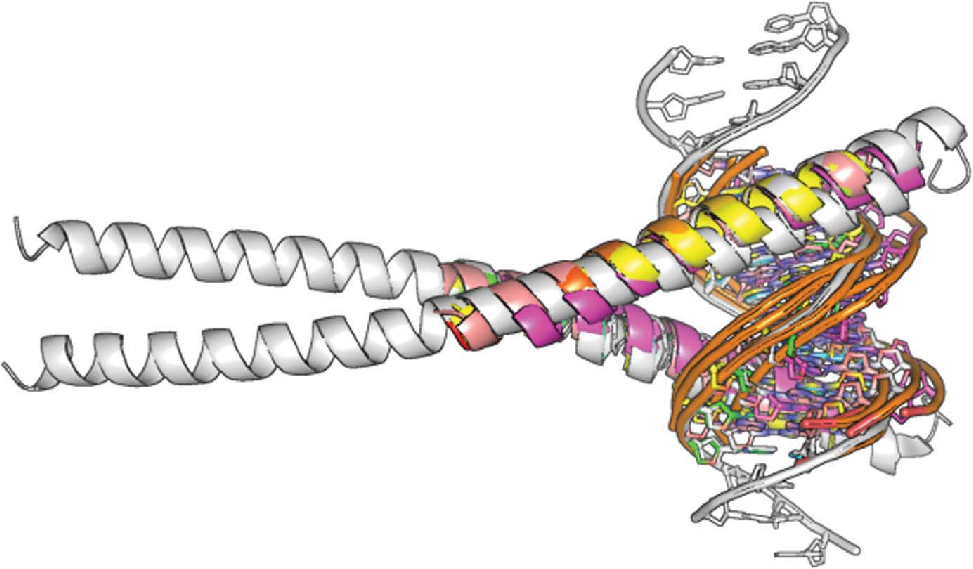
Zipper-type protein-DNA complex
ARCIMBOLDO on DNA: Structure solution of DNA-binding proteins and complexes with ARCIMBOLDO libraries
Kevin Pröpper#, Kathrin Meindl#, Massimo Sammito, Birger Dittrich, George M. Sheldrick, Ehmke Pohl* & Isabel Usón*.
Acta Cryst. D70, 1743-1757 (2014) (doi:10.1107/S1399004714007603)
# These authors contributed equally
* Corresponding authors