SHREDDER
SHREDDER: structure solution with ARCIMBOLDO using fragments derived from poor homology models
Improvement of search models against the experimental data, relying on PHASER’s rotation function and SHELXE pdb optimization
Molecular replacement relies on the availability of suitable models to be placed in the unit cell of the unknown structure in order to provide initial phases. ARCIMBOLDO uses small, very accurate fragments as search models. While it may not be evident which parts of a distant homolog will be most suitable purely from the degree of conservation, such a template is bound to contain building blocks, which might be further modified to enhance their accuracy.
ARCIMBOLDO-SHREDDER exploits fragments from distant homologs to phase macromolecular structures. Two modes are available within SHREDDER: spherical and sequential.
In SHREDDER_SPHERES, model generation uses three-dimensional volumes respecting structural units. The optimal fragment size is estimated from the expected log-likelihood gain (eLLG). Better sampling is attempted through model trimming or decomposition into rigid groups and optimization through Phaser’s gyre refinement. Also, after model translation, packing filtering and refinement, models are either disassembled into predetermined rigid groups and refined (gimble refinement) or PHASER’s LLG-guided pruning is used to trim the model of residues that are not contributing signal to the LLG at the target r.m.s.d. value. Phase combination among consistent partial solutions is performed in reciprocal space with ALIXE. Spherical shredding can improve models where deviations of the template from the target are evenly distributed among a similar fold.
In the SEQUENTIAL mode, the initial template is systematically shredded and fragments are scored against each unique solution of the rotation function. Results are combined into a score per residue and the template is trimmed accordingly. Models are selected by optimization of PHASER’s rotation function and trimming in SHELXE. Sequential shredding can improve models where the high average deviation from the target is owing to dissimilar or flexible regions.
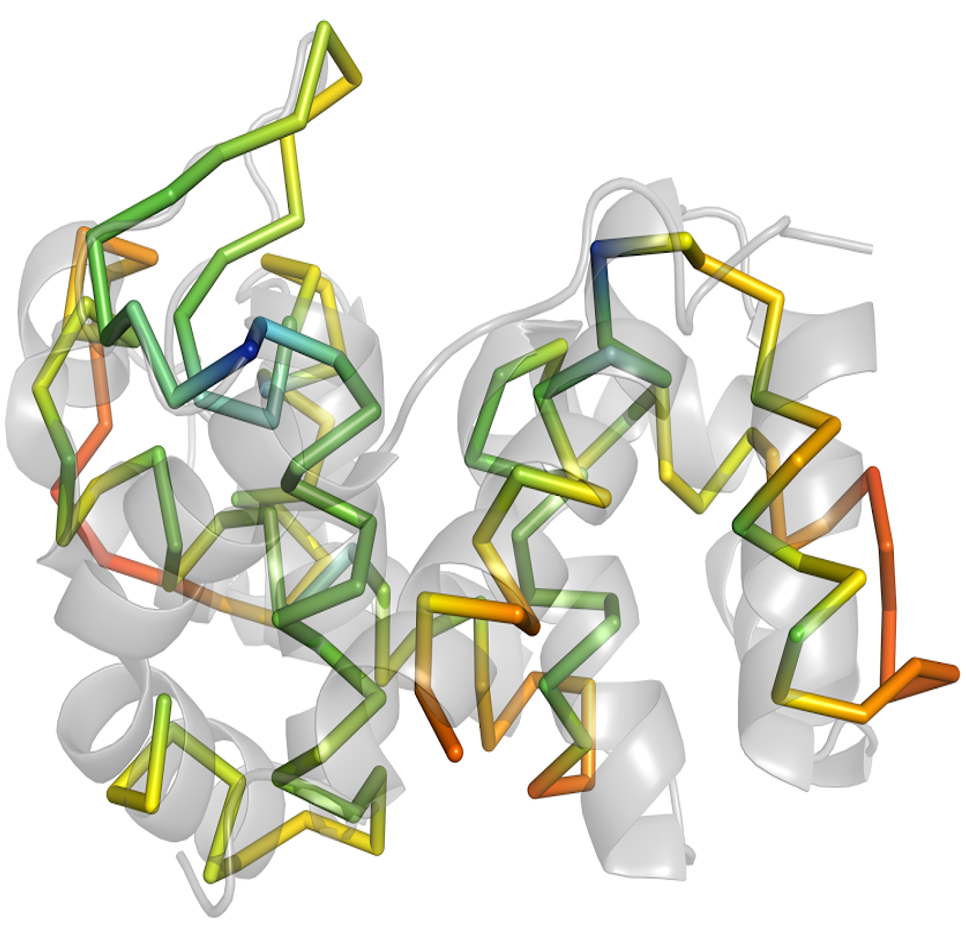
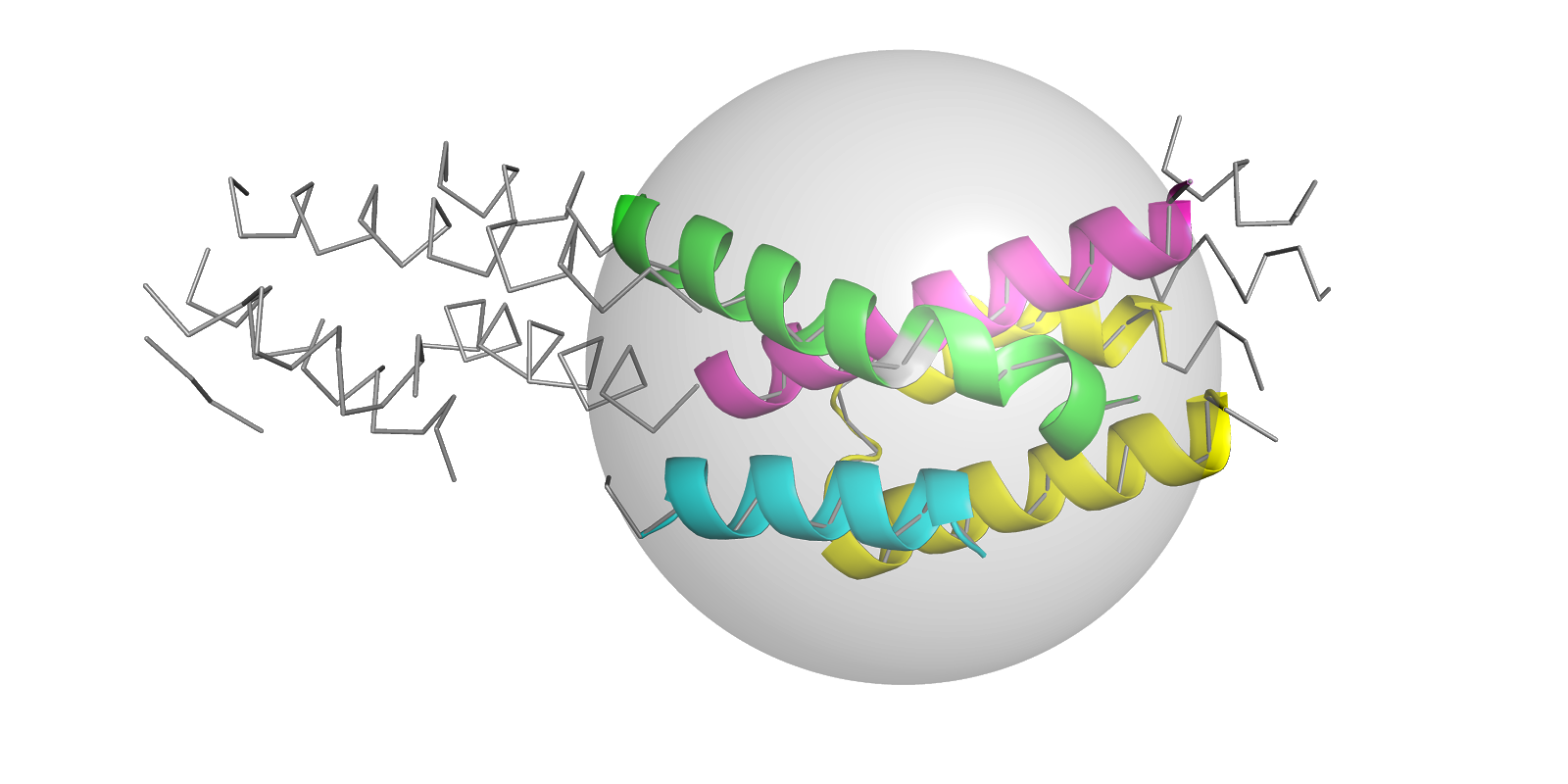
Structure solution with ARCIMBOLDO using fragments derived from distant homology models.
Massimo Sammito, Kathrin Meindl, Iñaki M. de Ilarduya, Claudia Millán, Cecilia Artola-Recolons, Juan A. Hermoso and Isabel Usón.
FEBS Journal 281: 4029–4045 (2014) (doi:10.1111/febs.12897)
Exploiting distant homologues for phasing through the generation of compact fragments, local fold refinement and partial solution combination.
Claudia Millán, Massimo Domenico, Airlie J. McCoy, Andrey F. Ziem Nascimento, Giovanna Petrillo, Robert D. Oeffner, Teresa Domínguez-Gil, Juan A. Hermoso, Randy J. Read and Isabel Usón
Acta Cryst. D74: 290-304 (2018) (doi:10.1107/S2059798318001365)